Research Interests
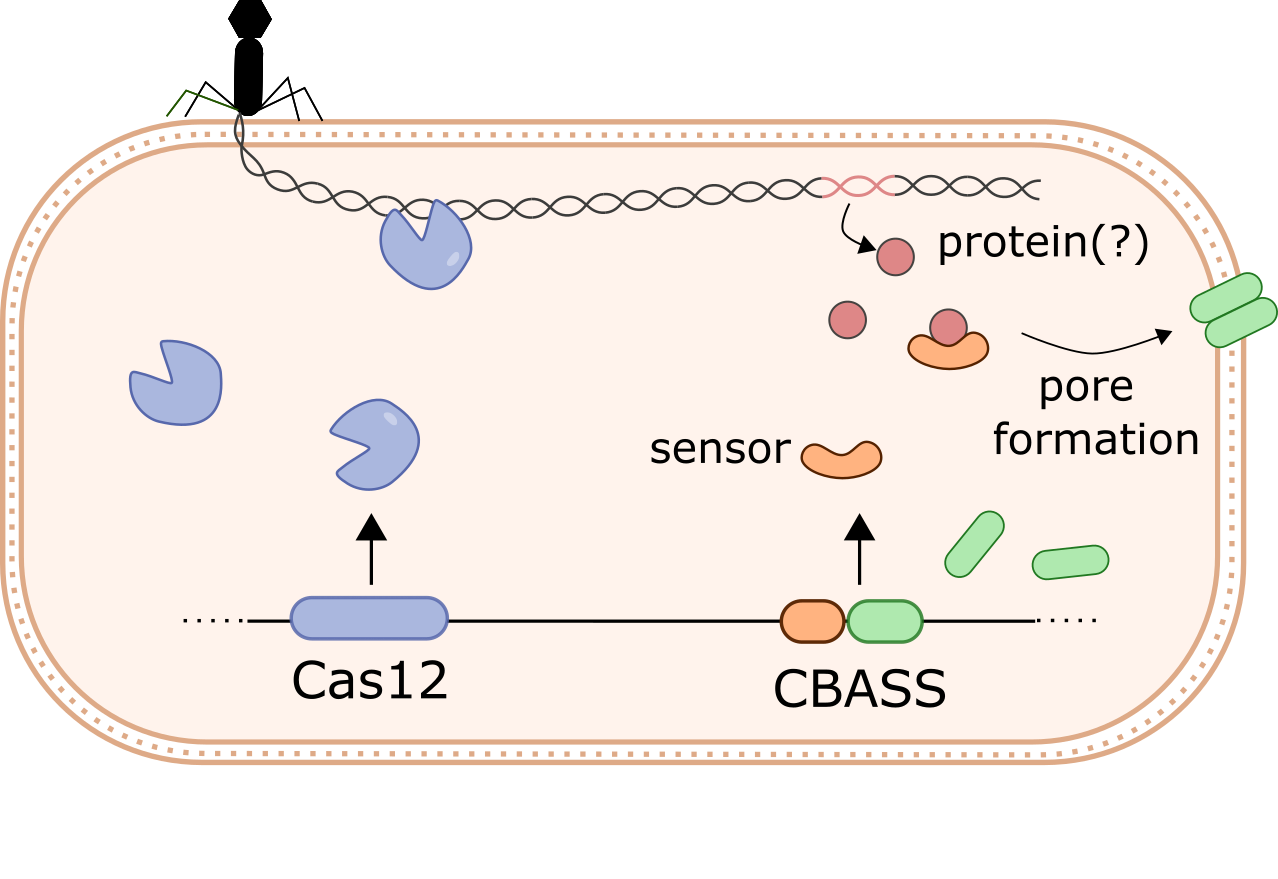
How do bacteria halt phage infection?
Dozens of new bacterial anti-phage systems have recently been found. Many of these systems are homologs of key eukaryotic immune proteins, such as cGAS and gasdermins, which appear to have originated in prokaryotes. While some anti-phage defense systems target phage directly (e.g. Cas12), others trigger cell death or dormancy to abort the infection and prevent phage spread. We are currently exploring these antiphage mechanisms using Klebsiella pneumoniae as a model organism.
How do phages overcome bacterial defenses?
Many bacteria encode multiple anti-phage systems, and closely related strains can differ dramatically in their defense repertoire. However, it is unclear how phages overcome these multi-layered defenses to replicate and spread throughout the population. We are currently exploring how phages overcome antiphage defenses in Klebsiella pneumoniae. We will use insights from this work to improve phage therapy for multidrug-resistant infections.
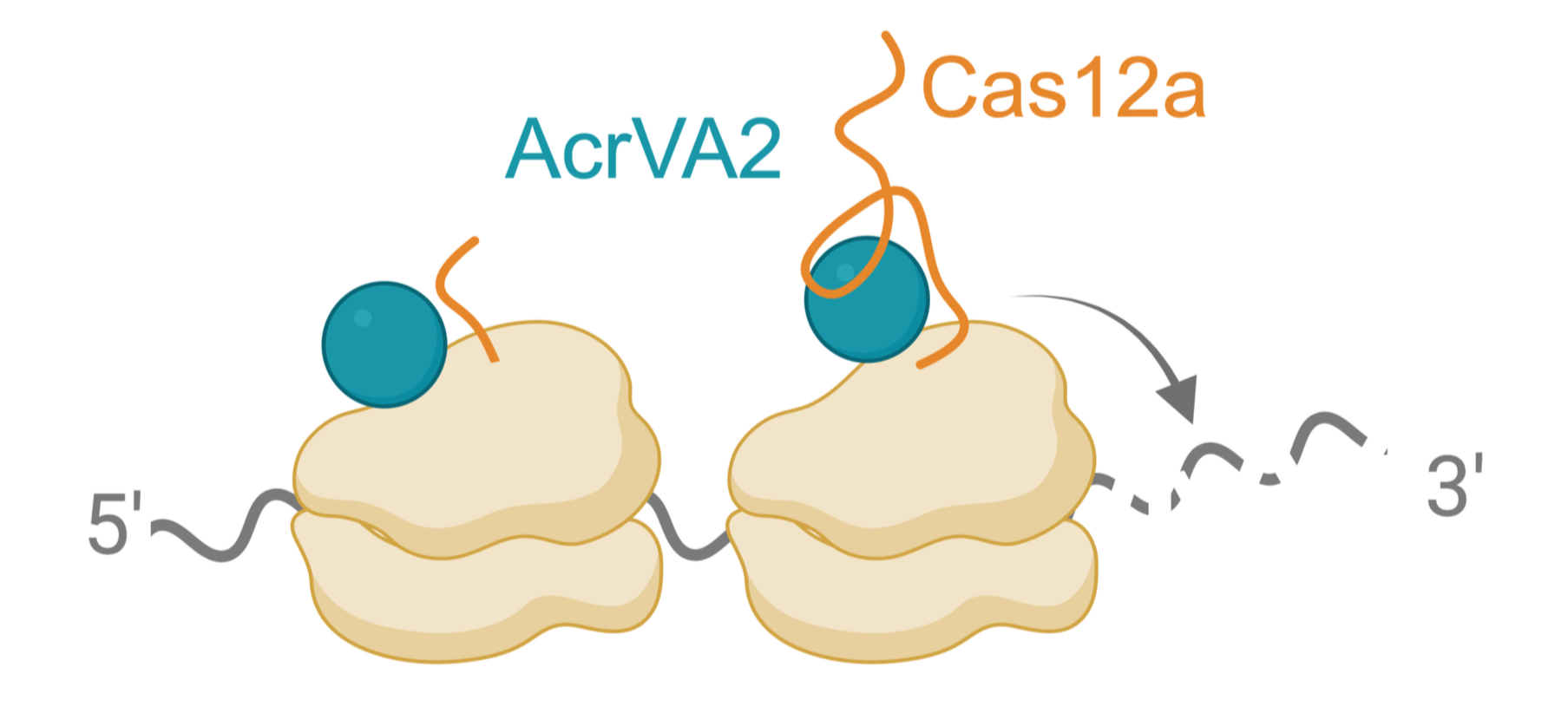
We discovered a phage-encoded anti-CRISPR (AcrVA2) that downregulates Cas12a by binding to its N-terminal polypeptide and triggering destruction of its mRNA during translation. AcrVA2 binds conserved and functionally important amino acids in Cas12a, allowing it to downregulate divergent Cas12a sequences while imposing a cost on Cas12a escape. This novel mechanism of molecular antagonism appears to be widespread against other genes in bacteria. In the future, we will explore this mechanism of polypeptide recognition and mRNA destruction while identifying other targets. We will also apply this mechanism to regulate expression of other genes in bacteria and beyond.
How does an anti-CRISPR trigger co-translational destruction of cas12a mRNA?